Moorhen Cryo-EM Tutorial: Fitting the Nanobody
Getting Started: Load the Map and Model
Get the reference files:
- https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/files/32143-partial-model.pdb
- https://ftp.ebi.ac.uk/pub/databases/emdb/structures/EMD-32143/map/emd_32143.map.gz
At the moment, you will need to gunzip the map.
- File → Coordinates →
32143-partial-model.pdb→ Open - File → CCP4/MRC Map → Choose File
emd_32143.map→ Open - Maps → Pull across the Radius to 77
-
(you can also use ‘[’ and ‘]’ to increase and decrease the map radius)
- Map Tools → Sharpen/Blur map
- B-factor to apply should be zero
- Turn on use resample and use the resample factor 1.4 → OK
- Maps → Click on the “Gear” icon on the (newly-created) Masked map → Draw settings → Activate “Lit lines”
You will see that this new map is more easy to interpret.
- Delete the first map:
- Maps Click on the “Gear icon” of the panel for the first map and Delete Map.
Rigid-Body Fitting
- Undisplay the new map:
- Click on the “Eye” icon
We are, at the moment, only interested in the parts of the map for which we don’t currently have a model. So now mask the map using the (partial) model (the parts of the model that we have already fitted):
- Map Tools → Map masking… → OK
You should now see a map with three unfitted parts
- the hormone
- a WD40 domain
- a nanobody domain
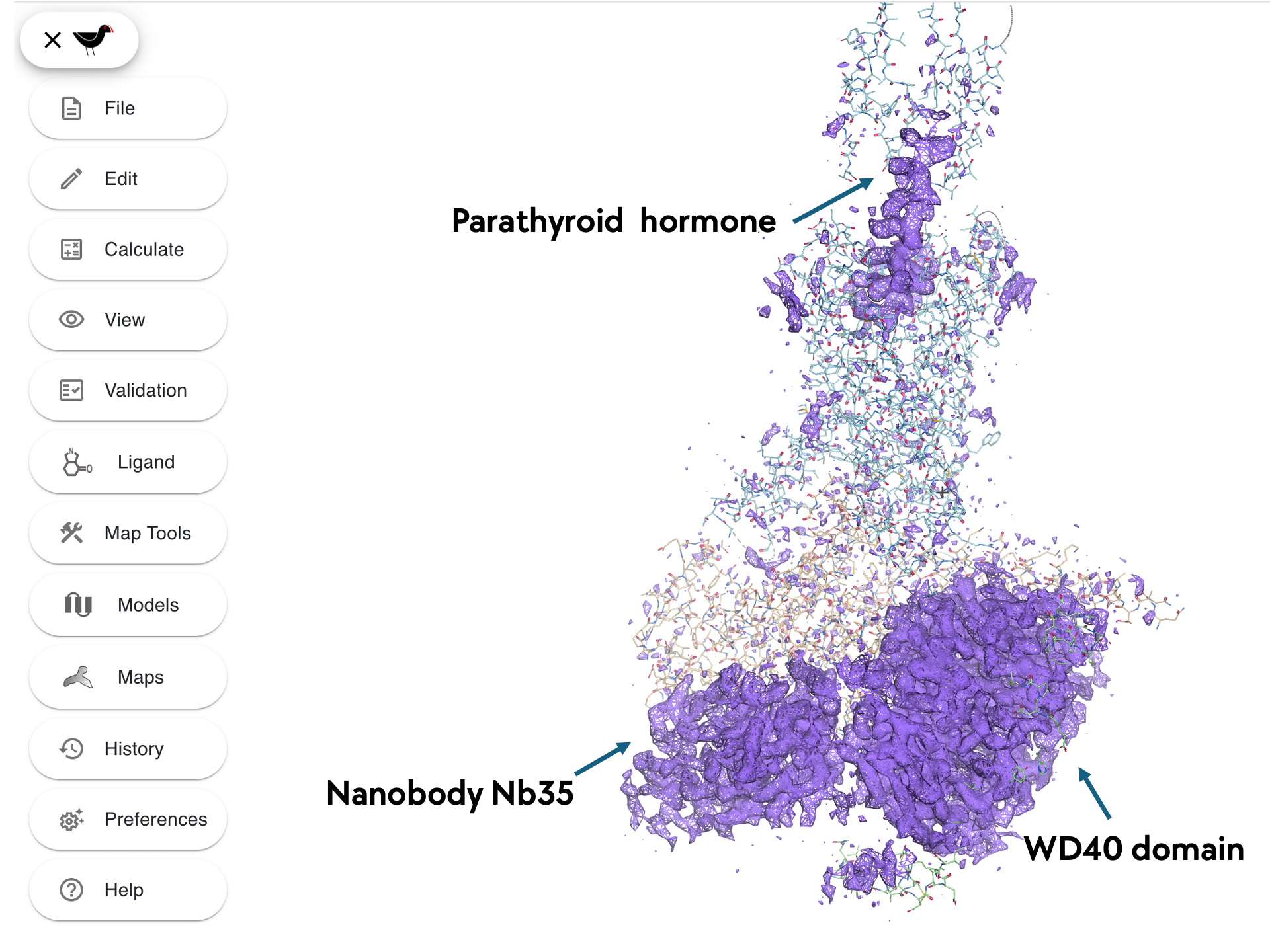
Fitting the Nanobody
We have done a BLAST search for the sequence of our nanobody and know that the structure was published already
in a different complex - it’s the “E” chain in the entry 6GDG. So let’s fetch that structure, extract
the “E” chain and then fit it to our reconstruction.
Fetch the Reference Structure:
- File → Fetch from Online Services →
6gdg
Now make the fragment we need from that:
- Edit → Copy Fragment
- Change “From molecule” to
6gdg - Selection to Copy:
//E - OK
So now we have the “E” chain floating around in space…
Delete the 6gdg model - we don’t need it any more:
- Models Click on the “Gear” icon of the
6gdgmolecule and Delete molecule
Change the representation of the ‘//E’ chain
- Models → Unclick the “Bonds” button and click the “Ribbons” button
Jiggle Fitting
Centre the view on the nanobody domain by “eyeballing” it and using middle-mouse click and drag. Rotate the view to check that you have found the approximate middle from various directions.
- Edit → Move Molecule Here →
- Select the molecule
6gdg fragment→ OK
Now to do the actual fitting:
- Calculate → Jiggle Fit with Fourier Filtering
- Choose molecule
6gdg fragment - Change the map to
Map 1 masked - Add an “Atom selection” as
//E - Change the number of trials to
310
Moorhen will think for a few seconds and then fit the nanobody into the density.
Refinement
- Change the ribbon representation back to bonds:
- Models → Unclick the “Ribbons” button and click the “Bonds” button
Now set-up the local distance restraints:
- Calculate → Generate Self Restraints
- Selection is “Molecule”
- Molecule is
6gdg fragment - Max Dist change to
5.1
Moorhen generates extra restraints (shown in grey lines)
- Maps → “Gear” icon of the masked map → set map weight
1830(it should be the default) → Set
To refine this domain/chain:
- Edit → Create a selection
- Molecule is
6gdg fragment - Atom Selection is
//(i.e. “all atoms”) - OK
You will see that the nanobody is now highlighted with green bonds and atoms. A dialog will appear at the middle top
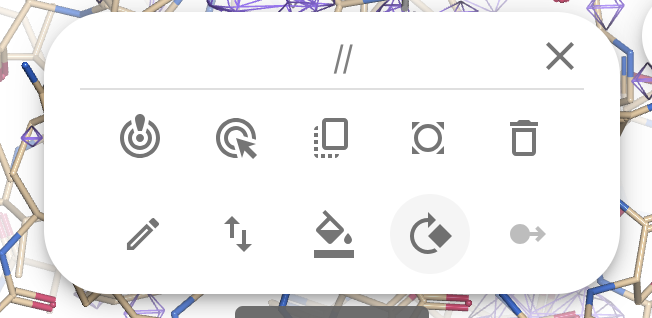
- Click on the “Refine” icon (top left)
- Moorhen refines the nanobody
- Accept the modification
Merge
After refinement we should merge the nanobody into the main molecule.
- Edit → Merge Molecues
- “From molecule” is
6gdg fragment - “Into molecule”
32143 partial model - OK
Now save the combined model:
- Models → Click on the “Download” icon of the
32143 partial model