Coot Goes Nuclear
I’ve recently updated Coot so that it preferentially uses nuclear distances for bond lengths.
The appropriate mmCIF tags to use are:
_chem_comp_bond.value_dist_nucleus and
_chem_comp_bond.value_dist_nucleus_esd
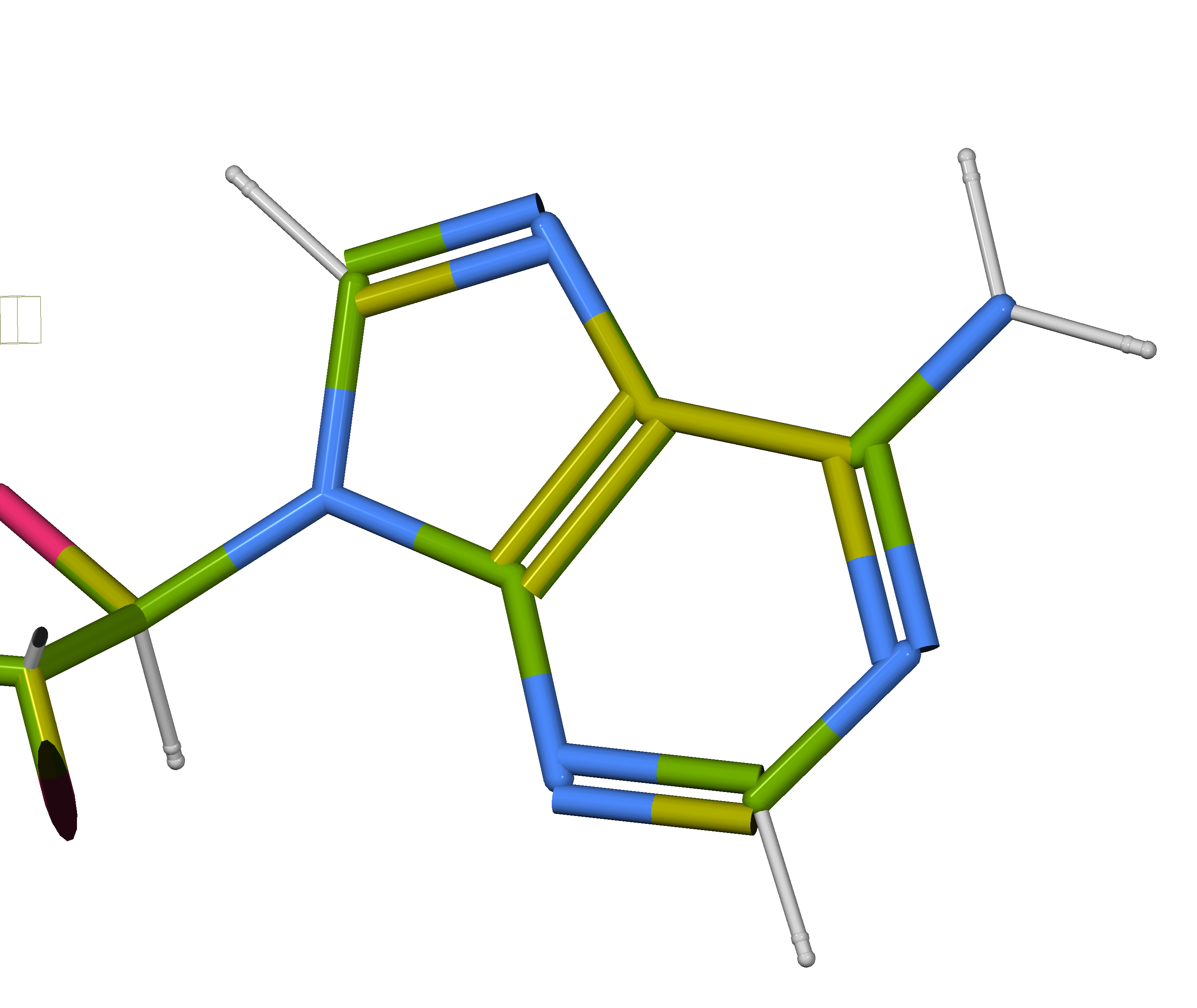
You can see here that the nuclear bond lengths are longer by 0.08 Å or so.
Additionally “Add Hydrogen Atoms” will add Hydrogen atoms using these bond lengths.
The longer Hydrogen atom bond lengths means different non-bonded contacts interactions during real space refinement. But there were problems. An analysis of the anomalies highlighted a few problem with incorrect non-bonded contact (for example, those of some types of Hydrogen bond). These were fixed and now the resulting model is now overall typically more consistent with Molprobity clash analysis:
$ molprobity.clashscore test.pdb keep_hydrogens=True nuclear=True
$ molprobity.molprobity test.pdb keep_hydrogens=True nuclear=True
This is available now in the refinement branch and will be available in 0.9.5.