Nature Front Cover Image
My colleagues recently published a paper (Nakane et al. (2020) “Single Particle cryo-EM at atomic resolution”) and asked me to make a figure. I did so and it is this week’s front cover image.
https://www.nature.com/nature/volumes/587/issues/7832
This was made with the “gtk3” branch of Coot.
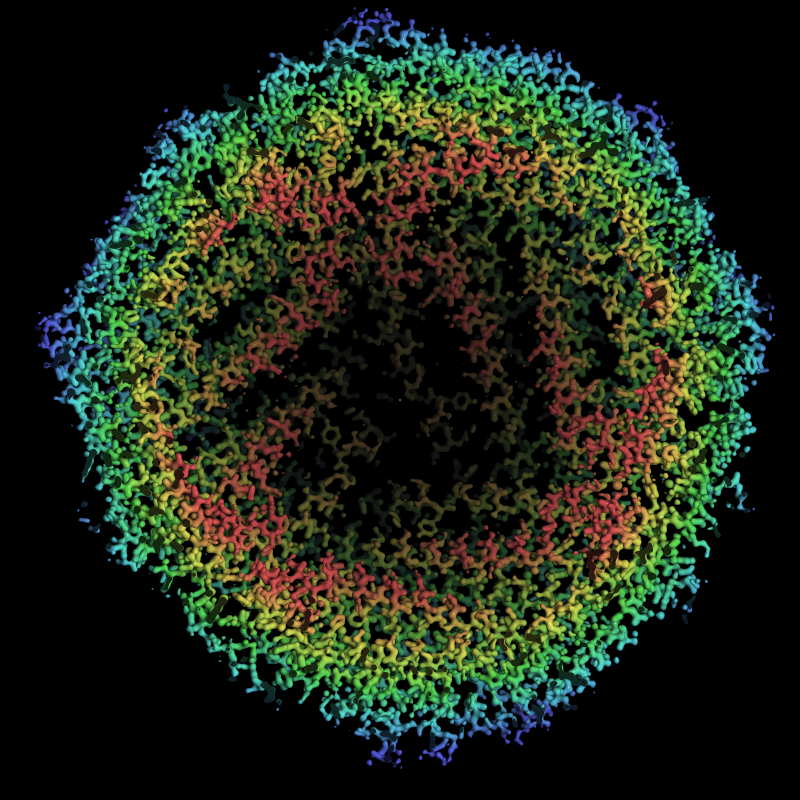
For radial colouring I used something like:
coot.set_draw_solid_density_surface(imol_map, 1)
coot.set_radial_map_colouring_min_radius(imol_map, 20)
coot.set_radial_map_colouring_max_radius(imol_map, 80)
coot.set_radial_map_colouring_invert(imol_map, 1)
coot.set_radial_map_colouring_enabled(imol_map, 1)Takanori himself made a front cover image too in PyMOL and seeing that I tried to make a version in a similar style using Coot. I didn’t copy it exactly, you can see that my version is not volumetric and is smoother and more gem-like.
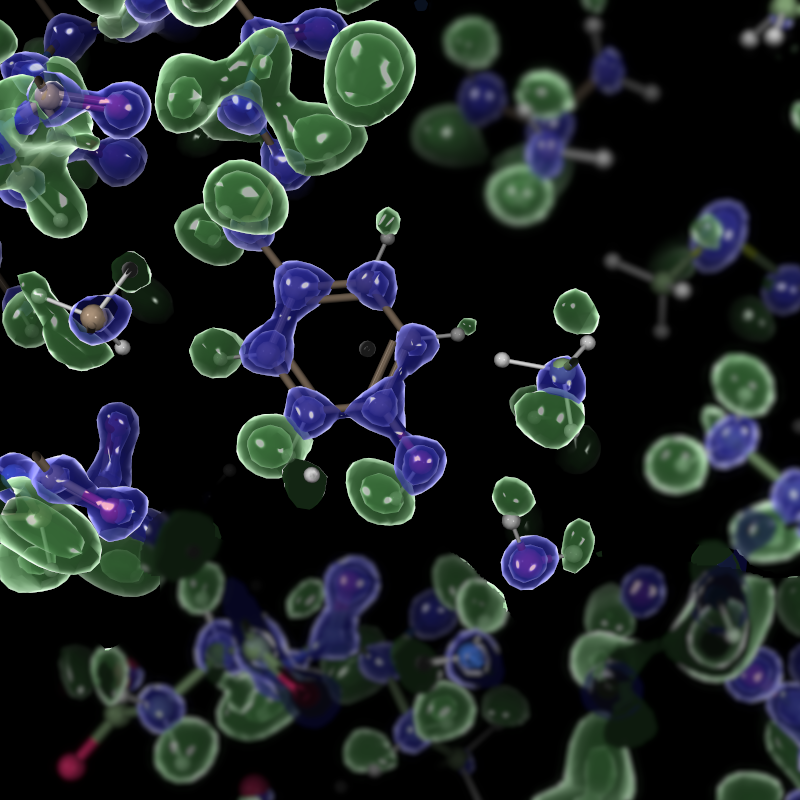
The point of this image is to illustrate that the reconstruction provides good evidence for the position of hydrogen atoms (which are the green blobs, of course).
High resolution version: This is intentionally large, to demonstrate the arbitrary framebuffer scaling.
You can see Takanori’s version on twitter (he is @biochem_fan)
The source code for “gtk3” Coot is updated/released every day or so, but the binaries are not (that’s too much of a time sink at the moment).
Here’s the script to make that second image:
import coot
b_factor = 1
resample_factor = 1.9
f1 = 0.0
coot.set_background_colour(f1,f1,f1)
coot.set_use_perspective_projection(1)
imol = read_pdb("ref1_n1_hydr_aniso_p-chainAL-HOH302.pdb")
coot.set_rotation_centre(65, 26, 32)
coot.set_bond_thickness(imol, 4)
imol_map = coot.handle_read_ccp4_map('apoF/aroundW302-normal.ccp4', 0)
imol_diff_map = coot.handle_read_ccp4_map('apoF/aroundW302-diff.ccp4', 0) # naughty
imol_map = coot.sharpen_blur_map_with_resampling(imol_map, b_factor, resample_factor);
imol_diff_map = coot.sharpen_blur_map_with_resampling(imol_diff_map, b_factor, resample_factor);
coot.set_draw_solid_density_surface(imol_map, 1)
coot.set_draw_solid_density_surface(imol_diff_map, 1)
coot.set_solid_density_surface_opacity(imol_map, 0.5);
coot.set_solid_density_surface_opacity(imol_diff_map, 0.5);
imol_map_copy = coot.copy_molecule(imol_map)
imol_diff_map_copy = coot.copy_molecule(imol_diff_map)
coot.set_map_colour(imol_map, 0.1, 0.1, 0.5)
coot.set_map_colour(imol_diff_map, 0.2, 0.4, 0.2)
coot.set_map_colour(imol_map_copy, 0.4, 0.4, 0.9)
coot.set_map_colour(imol_diff_map_copy, 0.4, 0.7, 0.4)
coot.set_draw_solid_density_surface(imol_map_copy, 1)
coot.set_draw_solid_density_surface(imol_diff_map_copy, 1)
coot.set_solid_density_surface_opacity(imol_map_copy, 0.15);
coot.set_solid_density_surface_opacity(imol_diff_map_copy, 0.15);
coot.set_map_material_specular(imol_map, 10, 100)
coot.set_map_material_specular(imol_diff_map, 10, 100)
coot.set_contour_level_absolute(imol_map, 0.135)
coot.set_contour_level_absolute(imol_map_copy, 0.208)
coot.set_contour_level_absolute(imol_diff_map, 0.155)
coot.set_contour_level_absolute(imol_diff_map_copy, 0.26)I applied the blur using the GIMP because Coot’s blur filter is not very good at the moment.