Making a Link with Coot and Acedrg
Coot makes use of the CCP4 Program “Acedrg” to make links between ligands and proteins.
Let’s try it out, first we will need to do a conventional ligand fitting procedure to import and fit the ligand.
Get the tutorial data:
http://www.ysbl.york.ac.uk/mxstat/JLigand/JLigand_link_tutorial.tar.gz
which is the “DATA” link in this page:
http://www.ysbl.york.ac.uk/mxstat/JLigand/tutorial_link.html
(We no longer recommend using JLigand for links, but the tutorial dataset is nice and useful)
Untar the file JLigand_link_tutorial.tar.gz - which gives you a directory called JLigand_link_tutorial
with files data.mtz and model.pdb
Let’s start up coot (having run the CCP4 set-up in that shell):
$ coot --pdb JLigand_link_tutorial/model.pdb
Now try to read in the data:
File → Open MTZ,mmcif,fcf or phs…
Then
“OK” in the following dialog
That runs REFMAC for us - takes 30 seconds so.
Display Manager
In the refmac-for-phases-tmp.pdb section, click the Display checkbutton to undisplay this model
Close (the Display Manager dialog)
Validate → Unmodelled Blobs → Find Blobs
Coot gives us a dialog - for me the blob of interest is “Blob 2” which is close to B258 LYS. Click Blob 2
When we are at the ligand site, you can dismiss the Blob dialog.
File → Get Monomer → PLP → OK
That should have imported a PLP, let’s undisplay it:
Display Manager → for the PLP_from_dist section, click the Display checkbutton to undisplay this model
Undisplay the difference map while you are there - it has served its purpose.
Close the Display Manager dialog
From the main menu bar: Ligand → Find Ligands
Click the “PLP_from_dict” checkbutton to select it
Where to Search? Choose “Right Here”
Find Ligands at the bottom of the dialog
OK the resulting Dialogs and see that the PLP has been fit reasonably well.
Let’s merge the new ligand into the protein molecule
Edit → Merge Molecules “5 Fitted ligand #0-0” → Merge (you may have a different molecule index)
Now the ligand is the same colour as the protein - indicating that the ligand has been merged.
Right…. so with that “preamble” done - we are ready to use Acedrg!
Calculate → Modules → CCP4, then
From the main menubar: CCP4 → Make Link via Acedrg
In the resulting dialog:
Order “Double”
On the formation of the link, an atom is removed from the ligand so entry in the Delete Atom entry “O4A” (without the quotes)
Start (Pick 2 Atoms)…
Click on the C4A of the ligand and the NZ of B 258 LYS.
Acedrg runs… it seems that nothing happens… we wait…
Then.. a dotted line has been formed between the NZ and the C4A indicating a LINK.
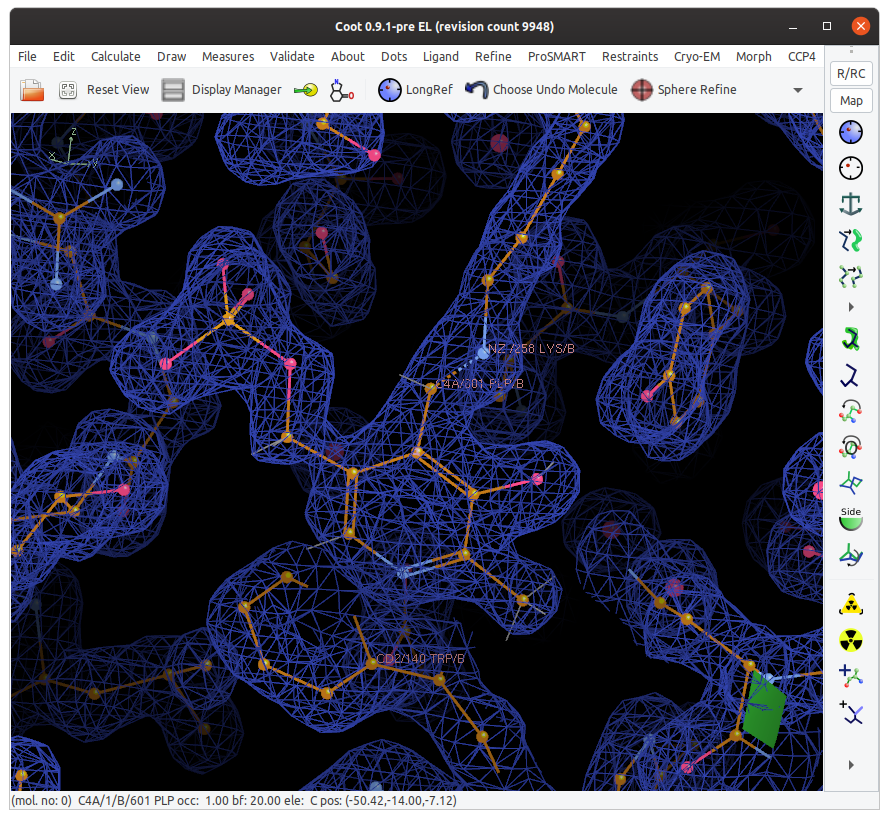
You can now run a “Sphere Refine” on that and the newly created link will be used in real space refinement.
In the terminal, you can see that the link cif file “acedrg-link-from-coot-PLP-LYS_link-hack.cif” has been written - if you wish to then import it to your refinement, say in CCP4i2.